This is the base class used to calculate the stomach content likelihood score based on the consumption of the preys in the model and the stomach content data specified in the input files. More...
#include <stomachcontent.h>
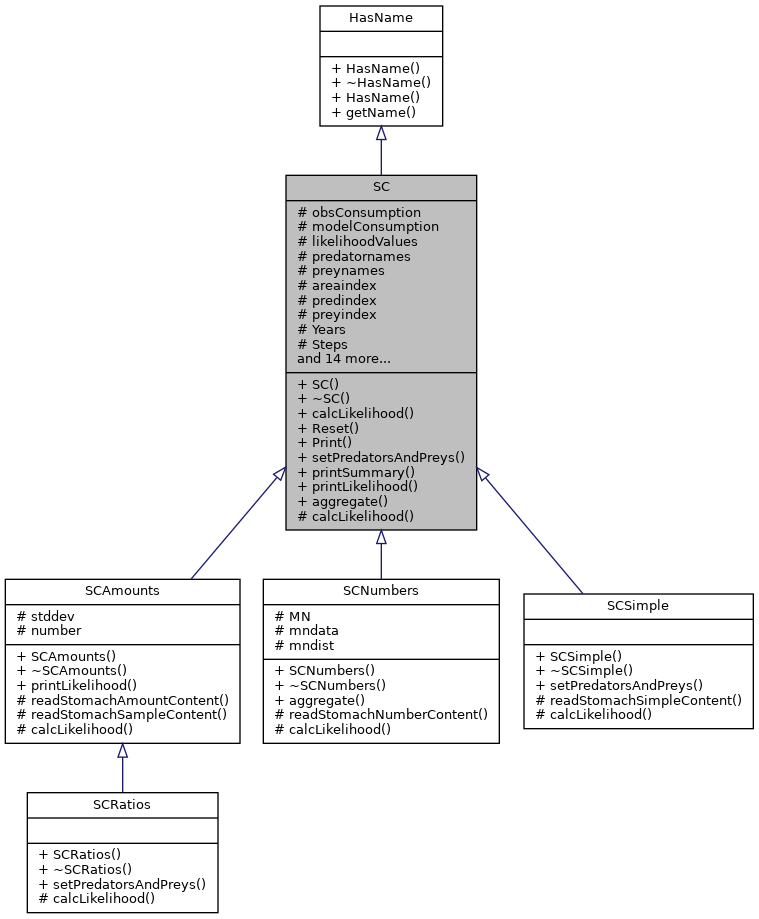
Public Member Functions | |
| SC (CommentStream &infile, const AreaClass *const Area, const TimeClass *const TimeInfo, Keeper *const keeper, const char *datafilename, const char *givenname) | |
| This is the SC constructor. More... | |
| virtual | ~SC () |
| This is the default SC destructor. More... | |
| virtual double | calcLikelihood (const TimeClass *const TimeInfo) |
| This function will calculate the likelihood score from the SC information. More... | |
| virtual void | Reset () |
| This function will reset the SC information. More... | |
| virtual void | Print (ofstream &outfile) const |
| This function will print the summary SC information. More... | |
| virtual void | setPredatorsAndPreys (PredatorPtrVector &Predators, PreyPtrVector &Preys) |
| This will select the predators and preys required to calculate the SC likelihood score. More... | |
| virtual void | printSummary (ofstream &outfile, double weight) |
| This function will print summary information from each StomachContent likelihood calculation. More... | |
| virtual void | printLikelihood (ofstream &outfile, const TimeClass *const TimeInfo) |
| This function will print information from each StomachContent calculation. More... | |
| virtual void | aggregate (int i) |
| This function will aggregate consumption information for each StomachContent calculation. More... | |
 Public Member Functions inherited from HasName Public Member Functions inherited from HasName | |
| HasName () | |
| This is the default HasName constructor. More... | |
| virtual | ~HasName () |
| This is the default HasName destructor. More... | |
| HasName (const char *givenname) | |
| This is the HasName constructor for a specified name. More... | |
| const char * | getName () const |
| This will return a null terminated text string containing the name of the object. More... | |
Protected Member Functions | |
| virtual double | calcLikelihood ()=0 |
| This function will calculate the likelihood score from the SC information. More... | |
Protected Attributes | |
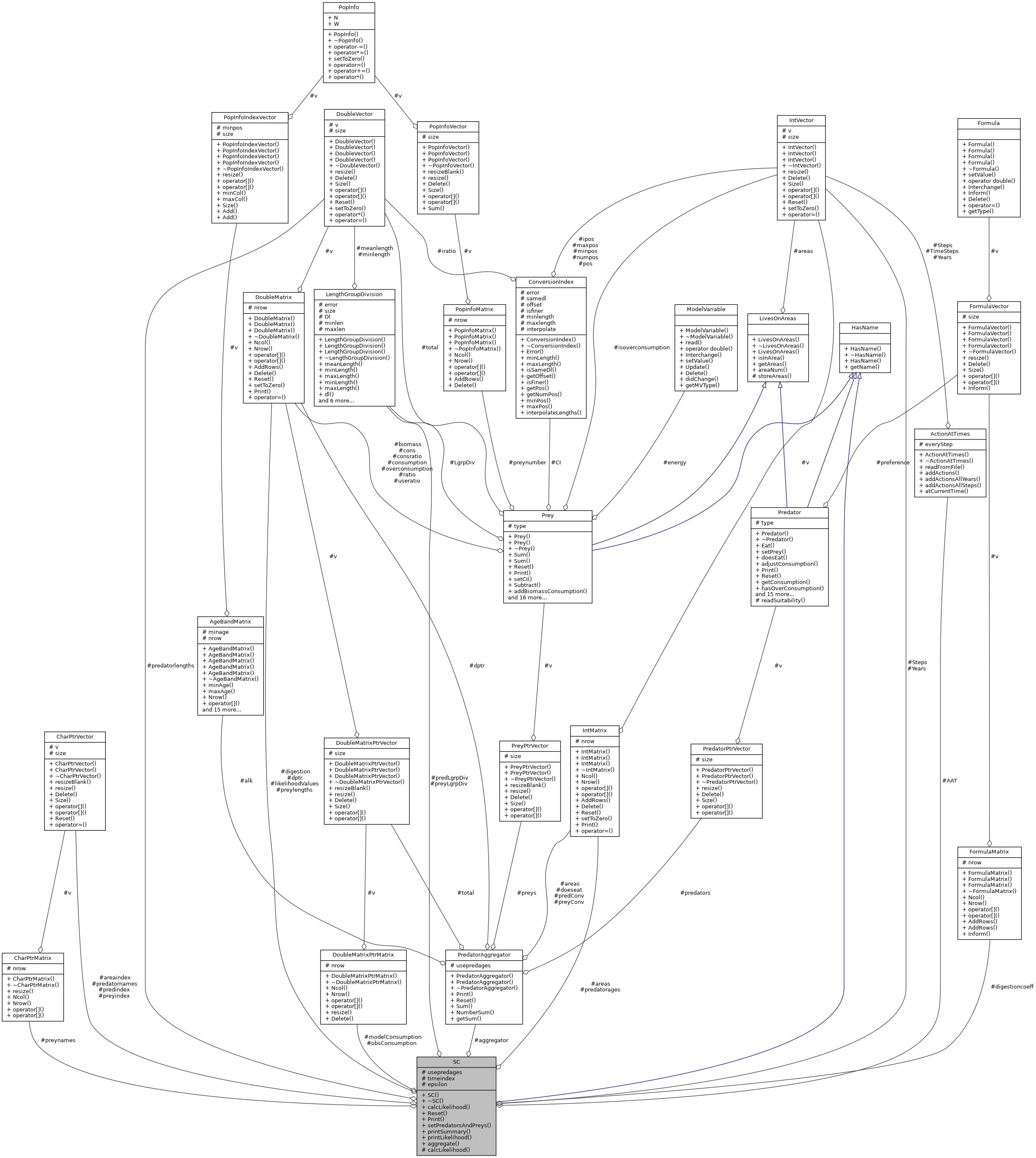
| DoubleMatrixPtrMatrix | obsConsumption |
| This is the DoubleMatrixPtrMatrix used to store stomach content (consumption) information specified in the input file. More... | |
| DoubleMatrixPtrMatrix | modelConsumption |
| This is the DoubleMatrixPtrMatrix used to store consumption (stomach content) information calculated in the model. More... | |
| DoubleMatrix | likelihoodValues |
| This is the DoubleMatrix used to store the calculated likelihood information. More... | |
| CharPtrVector | predatornames |
| This is the CharPtrVector of the names of the predators that will be used to calculate the likelihood score. More... | |
| CharPtrMatrix | preynames |
| This is the CharPtrMatrix of the names of the preys that will be used to calculate the likelihood score. More... | |
| CharPtrVector | areaindex |
| This is the CharPtrVector of the names of the areas. More... | |
| CharPtrVector | predindex |
| This is the CharPtrVector of the names of the predator aggregation units (either age or length) that will be read in from the input file. More... | |
| CharPtrVector | preyindex |
| This is the CharPtrVector of the names of the prey aggregation units that will be read in from the input file. More... | |
| IntVector | Years |
| This is the IntVector used to store information about the years when the likelihood score should be calculated. More... | |
| IntVector | Steps |
| This is the IntVector used to store information about the steps when the likelihood score should be calculated. More... | |
| DoubleVector | predatorlengths |
| This is the DoubleVector used to store predator length information. More... | |
| IntMatrix | predatorages |
| This is the IntMatrix used to store predator age information. More... | |
| DoubleMatrix | preylengths |
| This is the DoubleMatrix used to store prey length information. More... | |
| IntMatrix | areas |
| This is the IntMatrix used to store area information. More... | |
| int | usepredages |
| This is the flag used to denote whether the predators are specified using age groups or length groups. More... | |
| PredatorAggregator ** | aggregator |
| This is the PredatorAggregator used to collect information about the predation. More... | |
| LengthGroupDivision ** | preyLgrpDiv |
| This is the LengthGroupDivision used to store predator length information. More... | |
| LengthGroupDivision * | predLgrpDiv |
| This is the LengthGroupDivision used to store prey length information. More... | |
| int | timeindex |
| This is the index of the timesteps for the likelihood component data. More... | |
| ActionAtTimes | AAT |
| This ActionAtTimes stores information about when the likelihood score should be calculated. More... | |
| FormulaMatrix | digestioncoeff |
| This is the FormulaMatrix used to store the digestion parameters for the various predator prey pairs. More... | |
| DoubleMatrix | digestion |
| This is the DoubleMatrix used to store the calculated digestion coefficiants for the various predator prey pairs. More... | |
| double | epsilon |
| This is the value of epsilon used when calculating the likelihood score. More... | |
| const DoubleMatrix * | dptr |
| This is the DoubleMatrix used to temporarily store the information returned from the aggregatation function. More... | |
Detailed Description
This is the base class used to calculate the stomach content likelihood score based on the consumption of the preys in the model and the stomach content data specified in the input files.
- Note
- This will always be overridden by the derived classes that actually calculate the stomach content likelihood score
Constructor & Destructor Documentation
◆ SC()
| SC::SC | ( | CommentStream & | infile, |
| const AreaClass *const | Area, | ||
| const TimeClass *const | TimeInfo, | ||
| Keeper *const | keeper, | ||
| const char * | datafilename, | ||
| const char * | givenname | ||
| ) |
This is the SC constructor.
- Parameters
-
infile is the CommentStream to read the SC data from Area is the AreaClass for the current model TimeInfo is the TimeClass for the current model keeper is the Keeper for the current model datafilename is the name of the file containing the SC data givenname is the name for the likelihood component
◆ ~SC()
|
virtual |
This is the default SC destructor.
Member Function Documentation
◆ aggregate()
|
virtual |
This function will aggregate consumption information for each StomachContent calculation.
- Parameters
-
i is the index of the prey that is being consumed
Reimplemented in SCNumbers.
◆ calcLikelihood() [1/2]
|
protectedpure virtual |
◆ calcLikelihood() [2/2]
|
virtual |
◆ Print()
|
virtual |
This function will print the summary SC information.
- Parameters
-
outfile is the ofstream that all the model information gets sent to
◆ printLikelihood()
|
virtual |
This function will print information from each StomachContent calculation.
- Parameters
-
outfile is the ofstream that all the model likelihood information gets sent to TimeInfo is the TimeClass for the current model
Reimplemented in SCAmounts.
◆ printSummary()
|
virtual |
This function will print summary information from each StomachContent likelihood calculation.
- Parameters
-
outfile is the ofstream that all the model likelihood information gets sent to weight is the weight for the likelihood component
◆ Reset()
|
virtual |
This function will reset the SC information.
◆ setPredatorsAndPreys()
|
virtual |
This will select the predators and preys required to calculate the SC likelihood score.
- Parameters
-
Predators is the PredatorPtrVector of all the available predators Preys is the PreyPtrVector of all the available preys
Member Data Documentation
◆ AAT
|
protected |
This ActionAtTimes stores information about when the likelihood score should be calculated.
◆ aggregator
|
protected |
This is the PredatorAggregator used to collect information about the predation.
◆ areaindex
|
protected |
This is the CharPtrVector of the names of the areas.
◆ areas
◆ digestion
|
protected |
This is the DoubleMatrix used to store the calculated digestion coefficiants for the various predator prey pairs.
◆ digestioncoeff
|
protected |
This is the FormulaMatrix used to store the digestion parameters for the various predator prey pairs.
◆ dptr
|
protected |
This is the DoubleMatrix used to temporarily store the information returned from the aggregatation function.
◆ epsilon
|
protected |
This is the value of epsilon used when calculating the likelihood score.
◆ likelihoodValues
|
protected |
This is the DoubleMatrix used to store the calculated likelihood information.
- Note
- The indices for this object are [time][area]
◆ modelConsumption
|
protected |
This is the DoubleMatrixPtrMatrix used to store consumption (stomach content) information calculated in the model.
- Note
- The indices for this object are [time][area][predator][prey]
◆ obsConsumption
|
protected |
This is the DoubleMatrixPtrMatrix used to store stomach content (consumption) information specified in the input file.
- Note
- The indices for this object are [time][area][predator][prey]
◆ predatorages
◆ predatorlengths
|
protected |
This is the DoubleVector used to store predator length information.
◆ predatornames
|
protected |
This is the CharPtrVector of the names of the predators that will be used to calculate the likelihood score.
◆ predindex
|
protected |
This is the CharPtrVector of the names of the predator aggregation units (either age or length) that will be read in from the input file.
◆ predLgrpDiv
|
protected |
This is the LengthGroupDivision used to store prey length information.
◆ preyindex
|
protected |
This is the CharPtrVector of the names of the prey aggregation units that will be read in from the input file.
◆ preylengths
|
protected |
This is the DoubleMatrix used to store prey length information.
- Note
- The indices for this object are [prey number][prey length]
◆ preyLgrpDiv
|
protected |
This is the LengthGroupDivision used to store predator length information.
◆ preynames
|
protected |
This is the CharPtrMatrix of the names of the preys that will be used to calculate the likelihood score.
◆ Steps
|
protected |
This is the IntVector used to store information about the steps when the likelihood score should be calculated.
◆ timeindex
|
protected |
This is the index of the timesteps for the likelihood component data.
◆ usepredages
|
protected |
This is the flag used to denote whether the predators are specified using age groups or length groups.
◆ Years
|
protected |
This is the IntVector used to store information about the years when the likelihood score should be calculated.
The documentation for this class was generated from the following files:
- src/include/stomachcontent.h
- src/stomachcontent.cc